Week 9
Social Statistics and
Machine Learning
SOCI 316
Demystifying Regressions—
March 24th
Let’s Fit a Linear Probability Model
Caveat Emptor
Using a linear probability model (LPM) to predict a binary outcome can produce predicted probabilities—P(Y=1\mid X)—that fall outside the [0, 1] interval.
Let’s Fit a Linear Probability Model
We’re going to fit one anyway
(cf. von Hippel 2015).
Our Case
Our Task
To predict the probability of surviving the sinking of the Titanic as
a function of predictors like age, sex and passenger class.
P(\text{Survival} = 1 \mid X) = \beta_0 + X\beta + \varepsilon
A Simple Bivariate Model
Our Truncated Sample
Show the underlying code
# A tibble: 9 × 3
name survived age
<chr> <dbl> <dbl>
1 Aubart, Mme. Leontine Pauline 1 24
2 Stone, Mrs. George Nelson (Mart 1 62
3 Cardeza, Mr. Thomas Drake Marti 1 36
4 Buss, Miss. Kate 1 36
5 Smith, Miss. Marion Elsie 1 40
6 Hocking, Mr. Richard George 0 23
7 Cohen, Mr. Gurshon Gus 1 18
8 Cribb, Mr. John Hatfield 0 44
9 Skoog, Miss. Mabel 0 9Our First Model
P(\text{Survival} = 1 \mid X) = \beta_0 + \beta_1\text{age} + \varepsilon
We’re modelling the probability of survival as a function of age.
Our First Model
To fit this model, we need to calculate the slope and intercept parameters—that is, \beta_0 and \beta_1\text{age}, respectively.
Slopes can be derived using the following formula—
b = \frac{\sum{(X - \bar{X})(Y - \bar{Y})}}{\sum{(X - \bar{X})^2}}
Intercepts can be calculated as follows—
\alpha = \bar{Y} - b\bar{X}
Our First Model
Plugging in Our Values
| Y | X | X - \bar{X} | Y - \bar{Y} | (X - \bar{X})(Y - \bar{Y}) | (X - \bar{X})^2 |
|---|---|---|---|---|---|
| 1 | 24 | -8.44 | 0.33 | -2.81 | 71.31 |
| 1 | 62 | 29.56 | 0.33 | 9.85 | 873.53 |
| 1 | 36 | 3.56 | 0.33 | 1.19 | 12.64 |
| 1 | 36 | 3.56 | 0.33 | 1.19 | 12.64 |
| 1 | 40 | 7.56 | 0.33 | 2.52 | 57.09 |
| 0 | 23 | -9.44 | -0.67 | 6.33 | 89.14 |
| 1 | 18 | -14.44 | 0.33 | -4.81 | 208.53 |
| 0 | 44 | 11.56 | -0.67 | -7.74 | 133.64 |
| 0 | 9 | -23.44 | -0.67 | 15.67 | 549.64 |
| \sum (X - \bar{X}) = 0 | \sum (Y - \bar{Y}) = 0 | \sum (X - \bar{X})(Y - \bar{Y}) = 21.33 | \sum (X - \bar{X})^2 = 2008.92 |
\beta_1 = \frac{21.33}{2008.92} \approx 0.0106
\beta_0 = 0.667 - (0.0106 \times 32.44) \approx 0.3220
Our first LPM can therefore be summarized as follows—
\hat{P}(\text{Survived} = 1) = 0.3220 + 0.0106 \cdot \text{age} + \varepsilon
Our First Model
Plugging in Our Values
We can use programs like to streamline hand calculations.
Show the underlying code
library(tidyverse)
titanic_subset |> mutate(# Averages
mean_y = mean(survived),
mean_x = mean(age),
# To establish covariation between x and y
# Deviations from average, x
x_minus_mean_x = age - mean(age),
# Corresponding y value--deviations from average
y_minus_mean_y = survived - mean(mean_y),
# Tapping co-movement
slope_numerator = x_minus_mean_x * y_minus_mean_y,
# Squared residuals, positive values are yielded
slope_denominator = x_minus_mean_x^2) |>
# Putting it all together
mutate(# Summing up the values for the numerator ...
slope_numerator = sum(slope_numerator),
# And the denominator ...
slope_denominator = sum(slope_denominator),
# Yielding the slope estimate
slope = slope_numerator/slope_denominator,
# Which allows us to "solve" for the intercept:
intercept = mean_y - slope * mean_x) |>
select(intercept, slope) |>
distinct()# A tibble: 1 × 2
intercept slope
<dbl> <dbl>
1 0.322 0.0106Let’s Use Our Model—A Case Study
Kate Buss was 36 years old when the Titanic sank.
# A tibble: 1 × 3
name survived age
<chr> <dbl> <dbl>
1 Buss, Miss. Kate 1 36We can compute her probability of survival—using our clearly misspecified model—as follows:
Or we can use functions to generate these predictions for us.
Let’s Use Our Model
Range of Predictions—Truncated Sample
Show the underlying code
library(marginaleffects)
avg_predictions(survival_truncated, variables = "age") |>
as_tibble() |>
ggplot(mapping = aes(x = age, y = estimate)) +
geom_line(colour = "#b7a5d3", linewidth = 1.1) +
geom_ribbon(mapping = aes(ymin = conf.low,
ymax = conf.high),
fill = "lightgrey",
alpha = 0.3) +
theme_bw()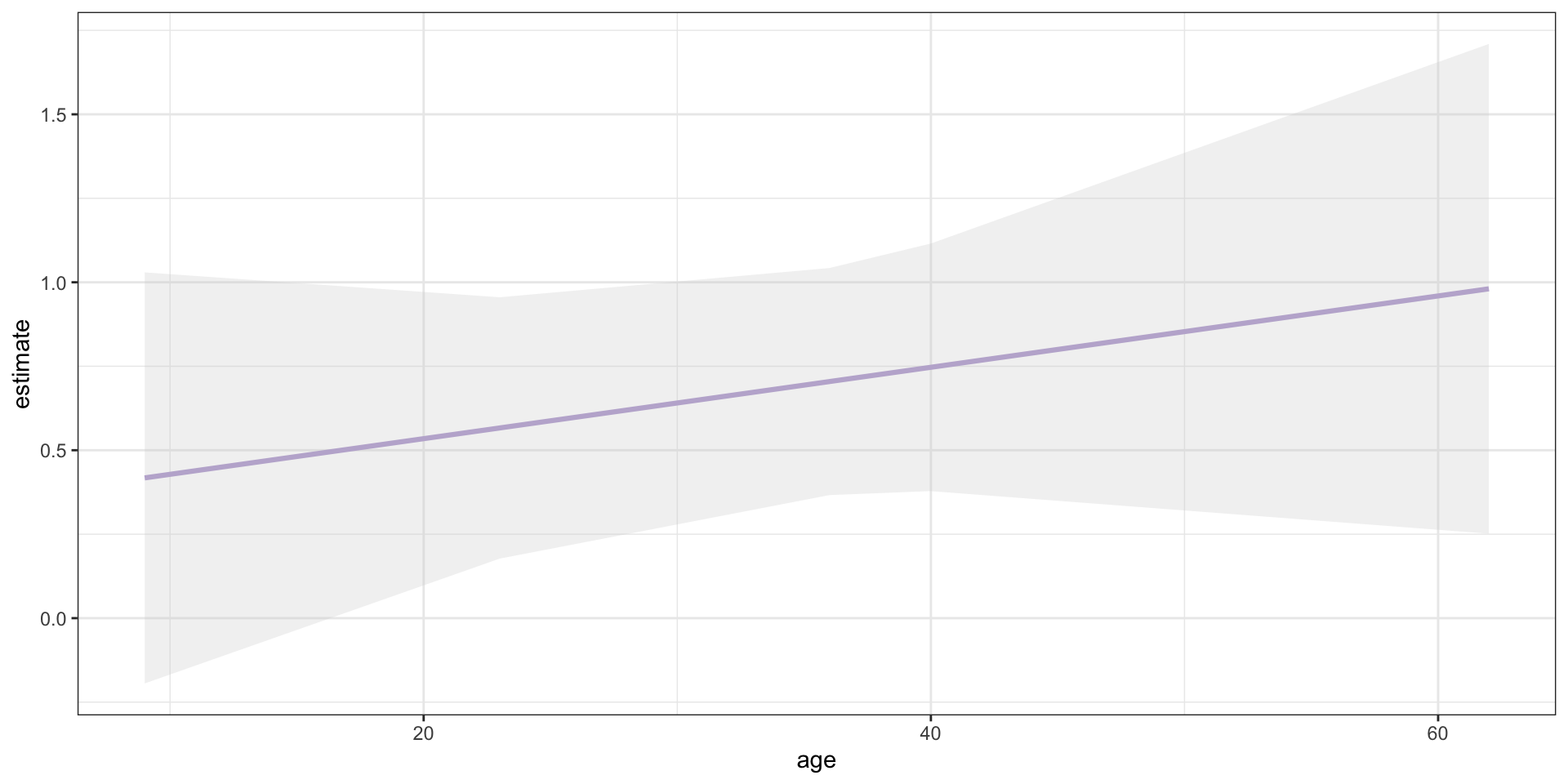
Let’s Use Our Model
Range of Predictions—Full Sample
Show the underlying code
survival_full <- lm(survived ~ age, data = titanic)
avg_predictions(survival_full, variables = "age") |>
as_tibble() |>
ggplot(mapping = aes(x = age, y = estimate)) +
geom_line(colour = "#b7a5d3", linewidth = 1.1) +
geom_ribbon(mapping = aes(ymin = conf.low,
ymax = conf.high),
fill = "lightgrey",
alpha = 0.3) +
theme_bw()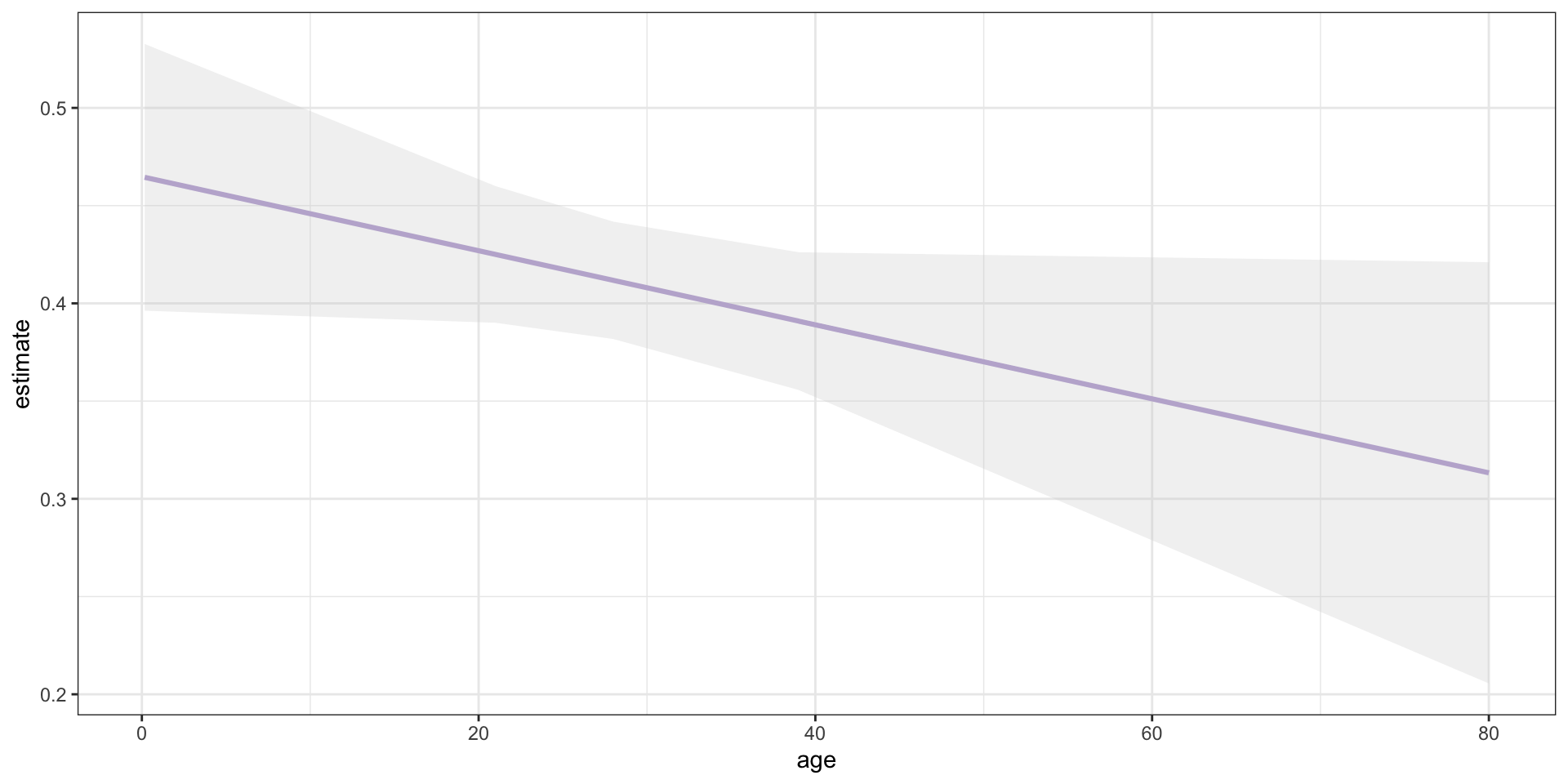
Let’s Use Our Model
Whoops!
Adding More Predictors
Updating the Full Model
Call:
lm(formula = survived ~ age + sex + passengerClass, data = titanic)
Residuals:
Min 1Q Median 3Q Max
-1.09442 -0.24408 -0.08375 0.23289 0.99397
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.1049549 0.0438210 25.215 < 2e-16 ***
age -0.0052695 0.0009316 -5.656 2.00e-08 ***
sexmale -0.4914131 0.0255520 -19.232 < 2e-16 ***
passengerClass2nd -0.2113738 0.0348568 -6.064 1.85e-09 ***
passengerClass3rd -0.3703874 0.0325039 -11.395 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.3913 on 1041 degrees of freedom
(263 observations deleted due to missingness)
Multiple R-squared: 0.3691, Adjusted R-squared: 0.3667
F-statistic: 152.3 on 4 and 1041 DF, p-value: < 2.2e-16Updating the Full Model
\begin{align*} \hat{P}(\text{Survived} = 1) = 1.105 - 0.00527 \cdot \text{age} - 0.4914 \cdot \text{sex}_\text{male} \\ - 0.2114 \cdot \text{class}_\text{2nd} - 0.3704 \cdot \text{class}_\text{3rd} + \varepsilon \end{align*}
Call:
lm(formula = survived ~ age + sex + passengerClass, data = titanic)
Coefficients:
(Intercept) age sexmale passengerClass2nd
1.104955 -0.005269 -0.491413 -0.211374
passengerClass3rd
-0.370387 We’re now modelling the probability of survival as a function of age, sex and passenger class.
Group Exercise I
Making Predictions
Here are three passengers selected at random:
# A tibble: 3 × 5
name survived sex age passengerClass
<chr> <dbl> <fct> <dbl> <fct>
1 Veal, Mr. James 0 male 40 2nd
2 Petersen, Mr. Marius 0 male 24 3rd
3 Johnson, Master. Harold Theodor 1 male 4 3rd In groups of 2-3, calculate the probability of survival for each passenger.
Call:
lm(formula = survived ~ age + sex + passengerClass, data = titanic)
Coefficients:
(Intercept) age sexmale passengerClass2nd
1.104955 -0.005269 -0.491413 -0.211374
passengerClass3rd
-0.370387 The “Solution”
Scroll for the code.
# A tibble: 3 × 2
name estimate
<chr> <dbl>
1 Veal, Mr. James 0.191
2 Petersen, Mr. Marius 0.117
3 Johnson, Master. Harold Theodor 0.222A Note About Uncertainty
Meaning in the Stars
===============================================
Dependent variable:
---------------------------
survived
-----------------------------------------------
age -0.005***
(0.001)
sexmale -0.491***
(0.026)
passengerClass2nd -0.211***
(0.035)
passengerClass3rd -0.370***
(0.033)
Constant 1.105***
(0.044)
-----------------------------------------------
Observations 1,046
R2 0.369
Adjusted R2 0.367
Residual Std. Error 0.391 (df = 1041)
F Statistic 152.257*** (df = 4; 1041)
===============================================
Note: *p<0.1; **p<0.05; ***p<0.01Standard Errors
SE(\hat{\beta}) = \sqrt{\frac{\hat{\sigma}^2}{\sum_{i = 1}^n (x_i- \bar{x})^2}}
Parameter estimates based on our full model.
# A tibble: 1 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
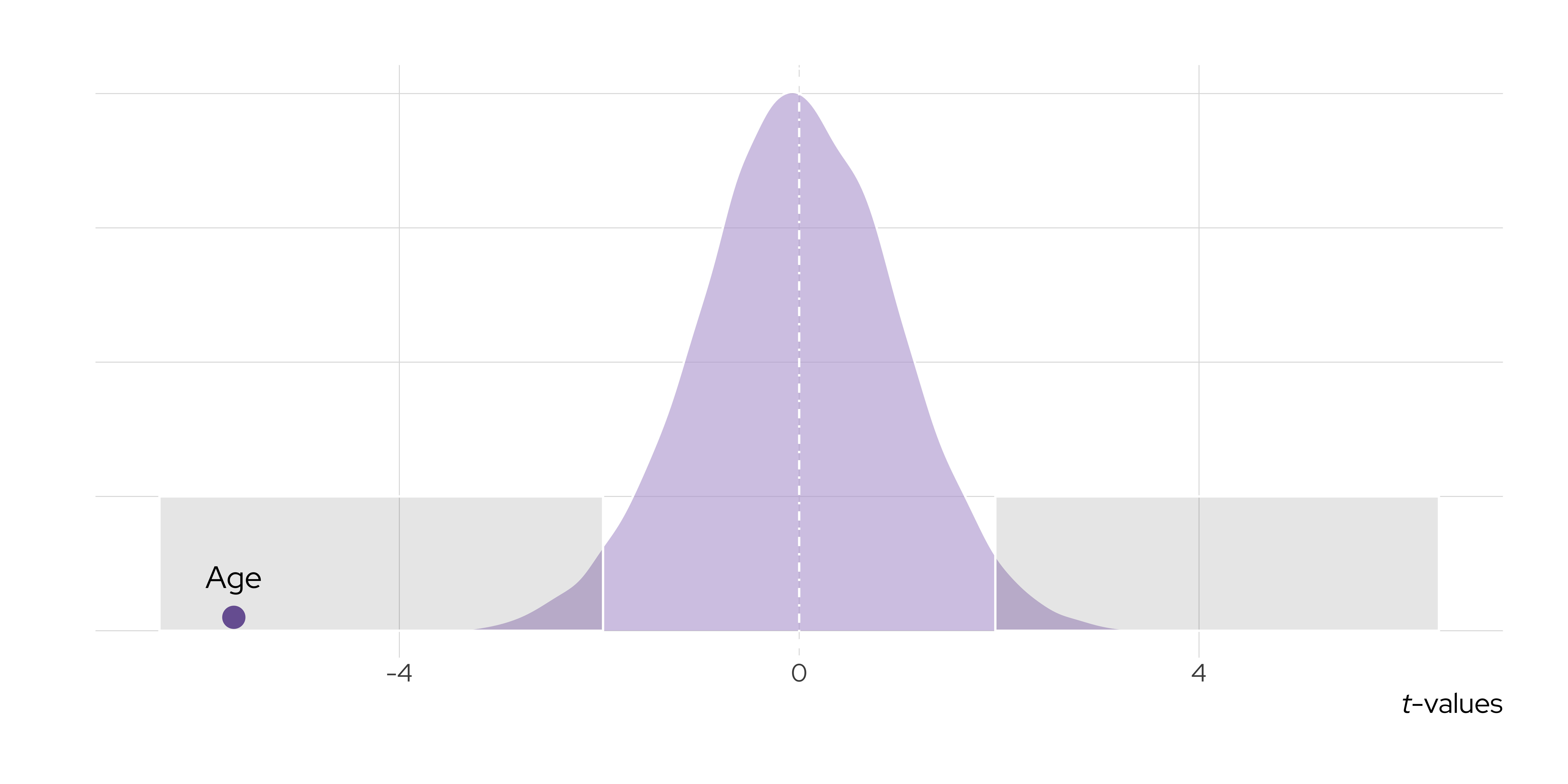
1 age -0.00527 0.000932 -5.66 0.0000000155Test Statistic Values
Hypothesis Testing
In Standard OLS Settings
t = \frac{\hat{\beta} - \color{856cb0}{\beta_{H_\theta}}}{SE(\hat{\beta})}
Hypothesis Testing
In Standard OLS Settings
t = \frac{\hat{\beta} - \color{856cb0}{\beta_{H_0}}}{SE(\hat{\beta})}
Show the underlying code
# A tibble: 1 × 6
term estimate std.error statistic manual_statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 age -0.00527 0.000932 -5.66 -5.66 0.0000000200Statistical Significance
According to Your Textbook
A result is statistically significant at the 5% level when the corresponding parameter is distinguishable from zero using 5% as the rejection threshold. Conversely, a result is not statistically significant at the 5% level when the corresponding parameter is not distinguishable from zero using 5% as the rejection threshold.
(Llaudet and Imai 2023, 215, EMPHASIS ADDED)
Memo II
Five Survey Items
Your next assignment is due this Friday at 8:00 PM. As a refresher—you are required to submit a short response memo proposing five potential survey questions.
For each question, you must include a \approx 100-word rationale explaining how the proposed item addresses a significant sociological question.
Please use the rest of today’s session to work on the memo!
A Gentle Introduction to Machine Learning—
March 27th
Informal Presentations
An Invitation
A Talk by Dr. Julian Go
Title
Policing Empires
Abstract
Click to Expand or Close
In the US and Britain, police bear the weapons and mindsets of armies, manifesting a “militarization” of policing. This lecture by Professor Julian Go offers a historical sociology of police militarization, revealing how, when and why it has occurred. It reveals what we call “police militarization” has been happening since the very founding of modern policing in the nineteenth century.
Date and Time
Location
What is Machine Learning?
Some Concrete Definitions
Grimmer and colleagues (2021, 396, EMPHASIS ADDED)
A class of flexible algorithmic and statistical techniques for prediction and dimension reduction.
Molina and Garip (2019, 28, EMPHASIS ADDED)
A way to learn from data and estimate complex functions that discover representations of some input (X), or link the input to an output (Y) in order to make predictions on new data.
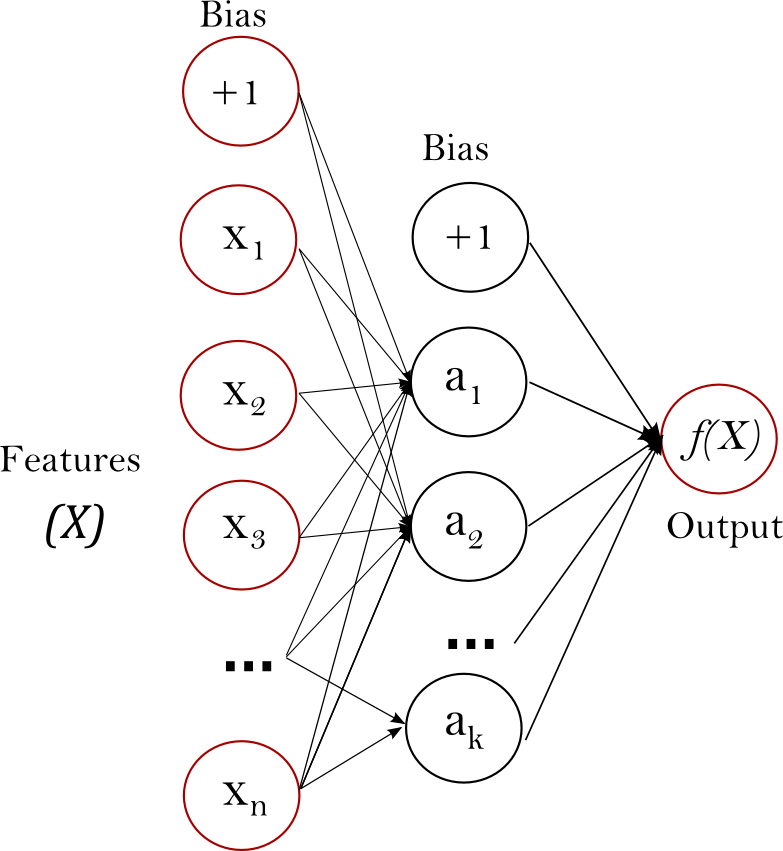
Supervised Machine Learning (SML)
High-Level Overview
Once deployed, SML algorithms learn the complex patterns linking X—a set of features (or independent variables)—to a target variable (or outcome), Y.
The goal of SML is to optimize predictions—i.e., to find functions or algorithms that offer substantial predictive power when confronted with new or unseen data.
Examples of SML algorithms include logistic regressions, random forests, ridge regressions, support vector machines and neural networks.
A quick note on terminology
If a target variable is quantitative, we are dealing with a regression problem.
If a target variable is qualitative, we are dealing with a classification problem.
Supervised Machine Learning (SML) cont.
An Illustration
# A tibble: 80,000 × 6
fruit shape weight_g colour texture origin
<chr> <chr> <dbl> <chr> <chr> <chr>
1 apple spherical 150 red crispy china
2 banana curved 120 yellow creamy ecuador
3 orange spherical 148 orange juicy egypt
4 watermelon spherical 4500 green juicy spain
5 strawberry conical 13 red juicy mexico
6 grape spherical 5 green juicy chile
7 mango ellipsoidal 240 yellow juicy india
8 pineapple conical 2100 yellow juicy costa rica
9 apple spherical 140 green crispy usa
10 banana curved 110 yellow creamy ecuador
# ℹ 79,990 more rows# A tibble: 1 × 6
fruit shape weight_g colour texture origin
<chr> <chr> <int> <chr> <chr> <chr>
1 <NA> conical 6 red juicy china # A tibble: 8 × 2
fruit probability
<chr> <dbl>
1 apple 0.03
2 banana 0
3 orange 0.05
4 watermelon 0
5 strawberry 0.71
6 grape 0.21
7 mango 0
8 pineapple 0 # A tibble: 1 × 1
prediction
<chr>
1 strawberry
Unsupervised Machine Learning (UML)
High-Level Overview
UML techniques search for a representation of the inputs (or features) that is more useful than X itself (Molina and Garip 2019).
- Put another way, UML algorithms search for hidden structure or latent patterns in high dimensional space.
In UML, there is no observed outcome variable Y—or target—to supervise the estimation process. Instead, we only have a vector of inputs to work with.
The goal in UML is to develop a lower-dimensional representation of complex data by inductively learning from the interrelationships among inputs.
- This can be achieved by reducing a vector of features to a smaller set of scales (e.g., via principal component analysis) or partitioning the sample into a small number of unobserved groups (e.g., via k-means clustering).
Unsupervised Machine Learning (UML) cont.
An Illustration
| V-Party Name | Label | Underlying Question |
|---|---|---|
v2paanteli |
Anti-Elitism | How important is anti-elite rhetoric for this party? |
v2papeople |
People-Centrism | Do leaders of this party glorify the ordinary people and identify themselves as part of them? |
v2paculsup |
Cultural Chauvinism | To what extent does the party leadership promote the cultural superiority of a specific social group or the nation as a whole? |
v2paminor |
Minority Rights | According to the leadership of this party, how often should the will of the majority be implemented even if doing so would violate the rights of minorities? |
v2paplur |
Political Pluralism | Prior to this election, to what extent was the leadership of this political party clearly committed to free and fair elections with multiple parties, freedom of speech, media, assembly and association? |
v2paopresp |
Demonization of Opponents | Prior to this election, have leaders of this party used severe personal attacks or tactics of demonization against their opponents? |
v2paviol |
Rejection of Political Violence | To what extent does the leadership of this party explicitly discourage the use of violence against domestic political opponents? |
Unsupervised Machine Learning (UML) cont.
An Illustration
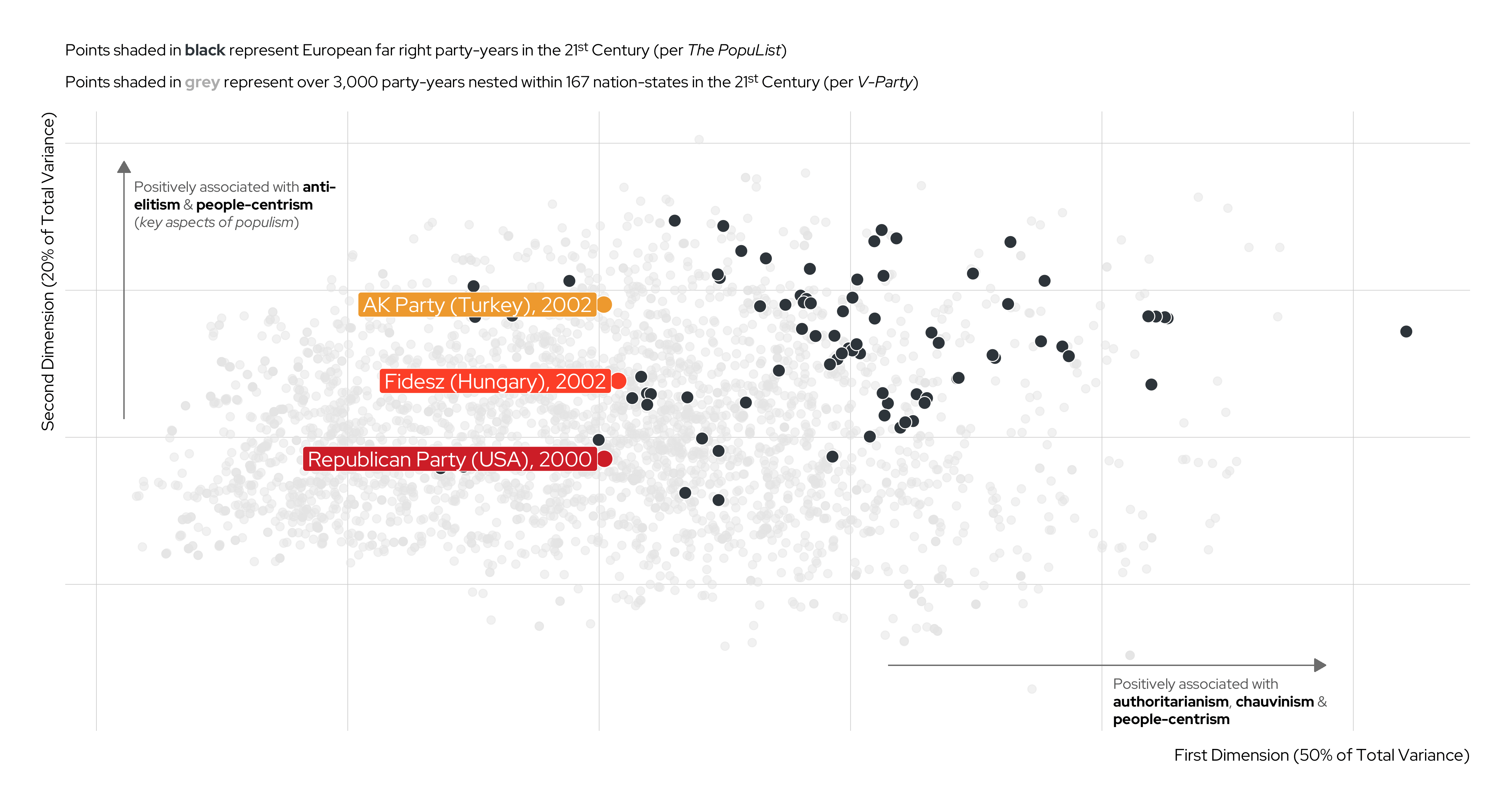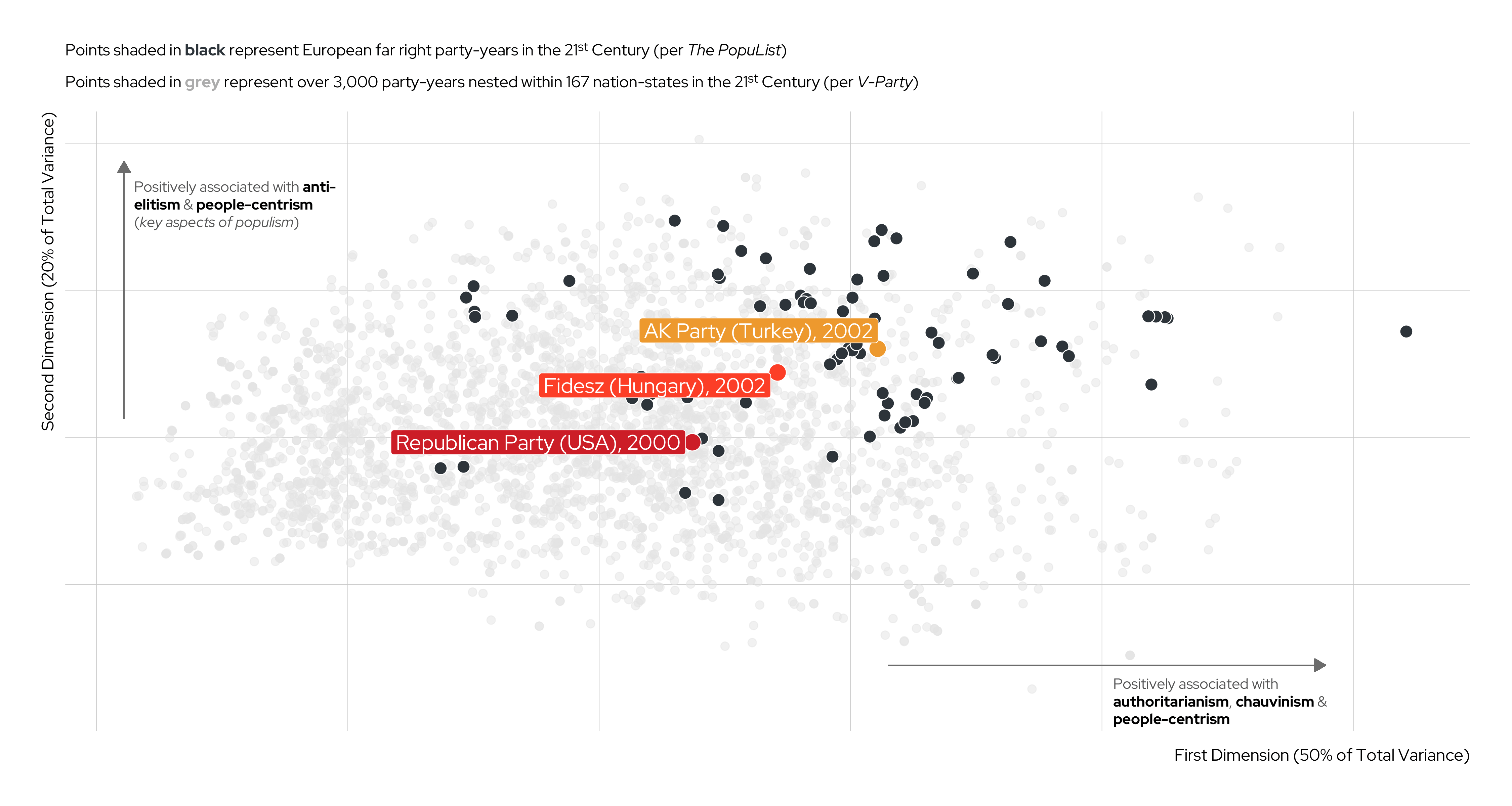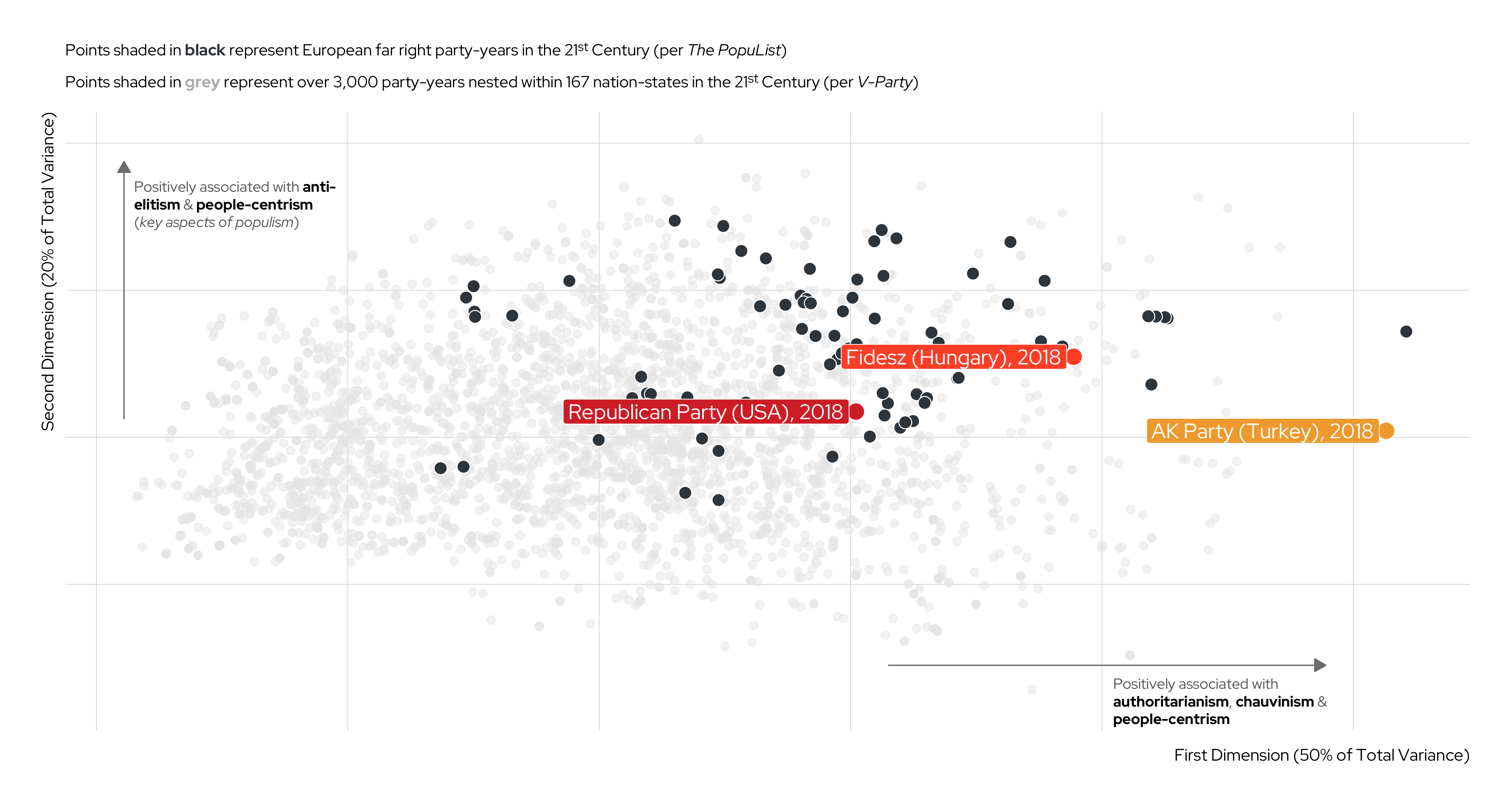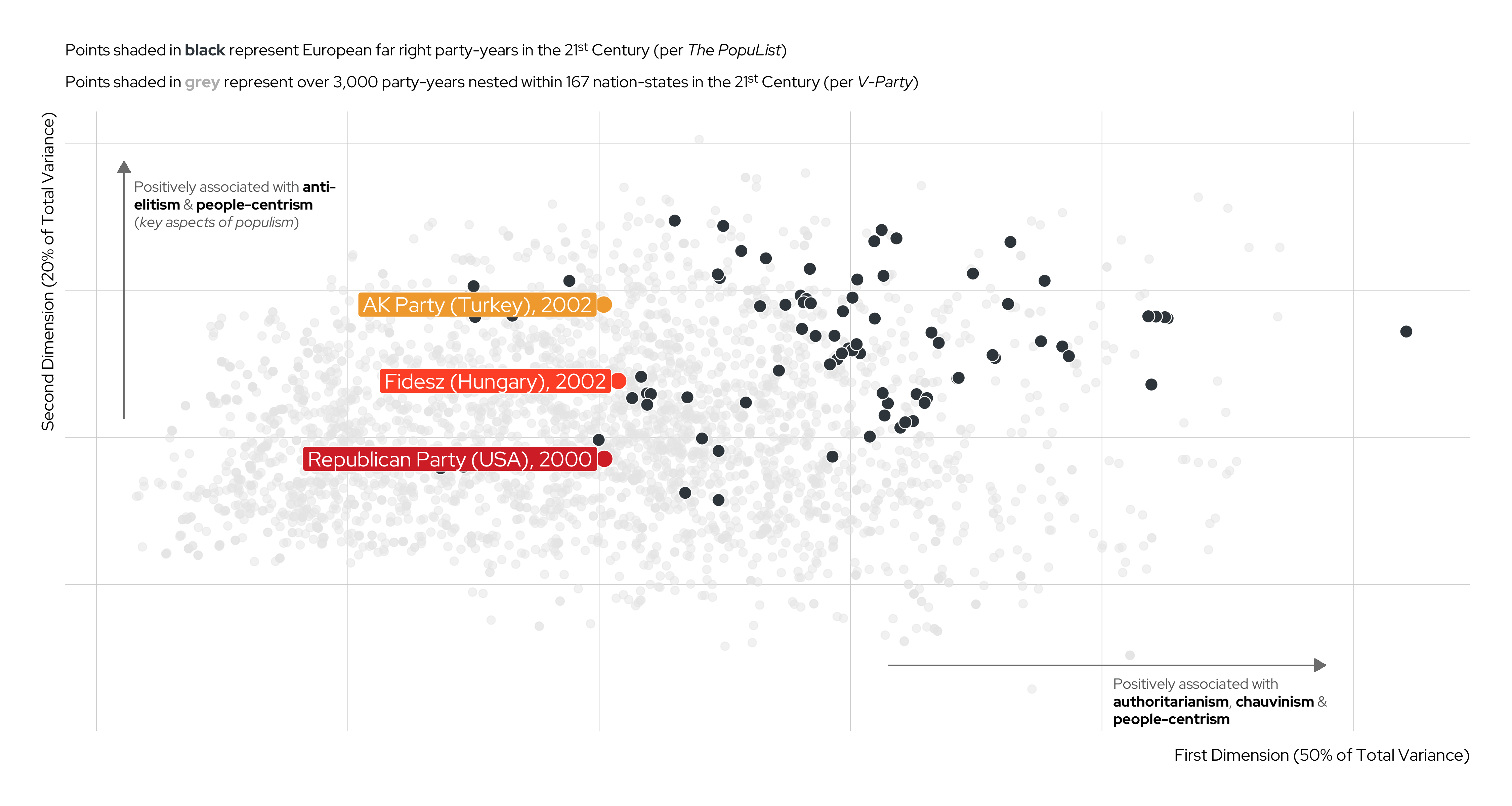
Karim and Lukk’s The Radicalization of Mainstream Parties in the 21st Century
Machine Learning vs Classical Parametric Statistics
The Two Cultures
As Grimmer and colleagues (2021) note, “machine learning is as much a culture defined by a distinct set of values and tools as it is a set of algorithms.”
This point has, of course, been made elsewhere.
Leo Breiman (2001) famously used the imagery of warring cultures to describe two major traditions—(i) the generative modelling culture and (ii) the predictive modelling culture—that have achieved hegemony within the world of statistical modelling.
The terms generative and predictive (as opposed to data and algorithmic) come from David Donoho’s (2017) 50 Years of Data Science.
The Two Cultures cont.
| Quantity of Interest | Primary Goals | Key Strengths | Key Limitations |
|---|---|---|---|
| Generative (i.e., Classical Statistics) | |||
| Inferring relationships between X and Y | Interpretability; emphasis on uncertainty around estimates; explanatory power | Bounded by statistical assumptions, inattention to variance across samples | |
| Predictive (i.e., Machine Learning) | |||
| Generating accurate predictions of Y | Predictive power; potential to simplify high dimensional data; relatively unconstrained by statistical assumptions | Inattention to explanatory processes, opaque links between X and Y | |
Note: To be sure, the putative strengths and weaknesses of these modelling “cultures” have been hotly debated.
The Affordances of Machine Learning
Advances in machine learning can provide empirical leverage to social scientists and sharpen social theory in one fell swoop.
Lundberg, Brand and Jeon (2022), for instance, argue that adopting a machine learning framework can help social scientists:
- Amplify human coding
- Summarize complex data structures
- Relax statistical assumptions
- Target researcher attention
While ML is often associated with induction, van Loon (2022) argues that SML algorithms can help us deductively resolve predictability hypotheses as well.
- What plays a larger role in shaping human behaviour—nature or nurture?
Key Terms and Concepts in the SML Setting
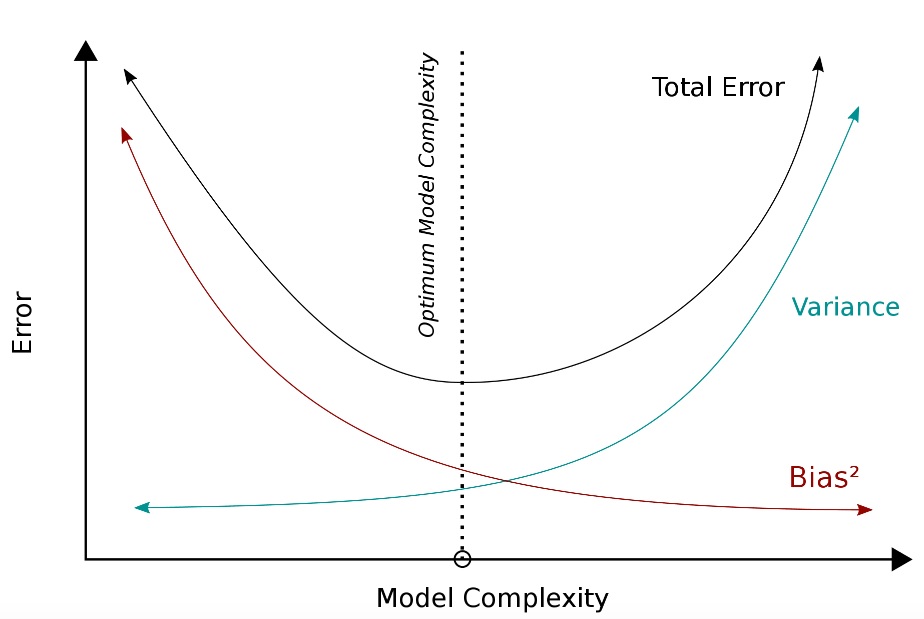
Bias-Variance Tradeoff
Image can be retrieved here.
Bias emerges when we build SML algorithms that fail to sufficiently map the patterns—or pick up the empirical signal–linking X and Y. Think: underfitting.
Variance arises when our algorithms not only pick up the signal linking X and Y, but some of the noise in our data as well. Think: overfitting.
When adopting an SML framework, researchers try to strike the optimal balance between bias and variance.
Training, Validation and Testing
- In an SML setting, we want to reduce our algorithm’s generalization or test error—i.e., “the prediction error of a model on new data” (Molina and Garip 2019).
- To arrive at an estimate of our model’s performance, we can (randomly) partition our global sample of observations into disjoint sets or subsamples.
We can use a training set to fit our algorithm—to find weights (or coefficients), recursively split the feature space to grow decision trees and so on.
Training data should constitute the largest of our three disjoint sets.
We can use a validation set to find the right estimator out of a series of candidate algorithms—or select the best-fitting parameterization of a single algorithm.
Often, using both training and validation sets can be costly: data sparsity can give rise to bias.
Thus, when limited to smaller samples, analysts often combine training and validation—say, by recycling training data for model tuning and selection.
We can use a testing set to generate a measure of our model’s predictive accuracy (e.g., the
F1 scorefor classification problems)—or to derive our generalization error.This subsample is used only once (to report the performance metric); put another way, it cannot be used to train, tune or select our algorithm.
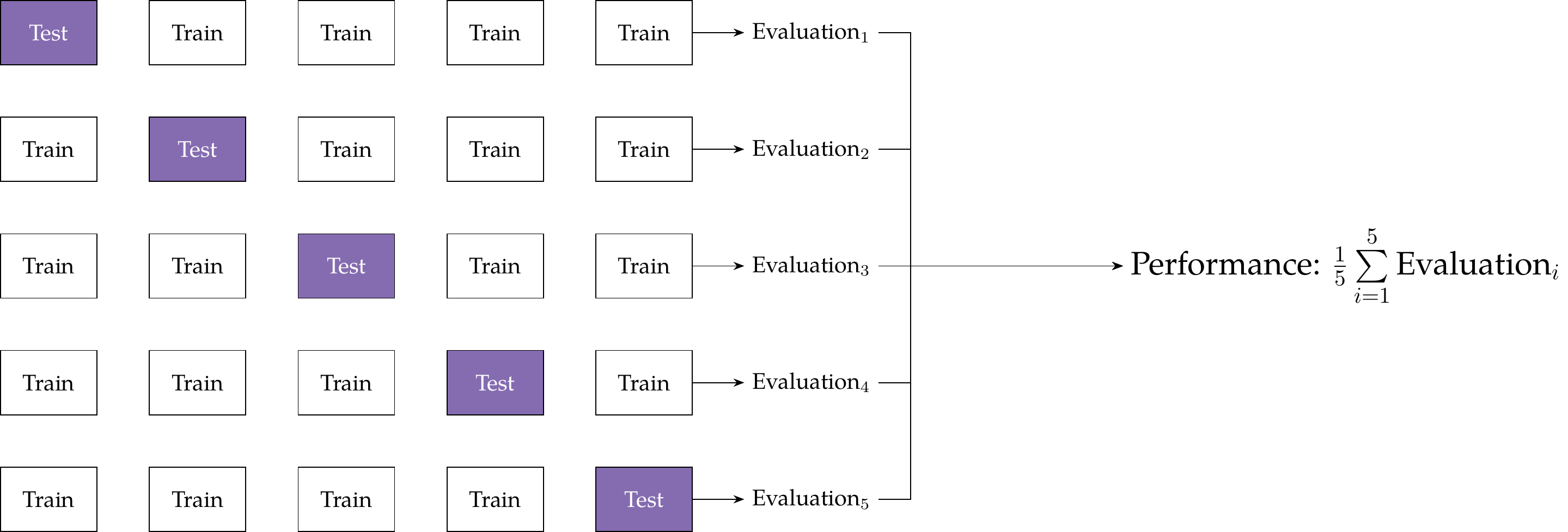
k-Fold Cross-Validation
Unlike conventional approaches to sample partition, k or v-fold cross-validation allows us to learn from all our data.
k-fold cross-validation proceeds as follows:
- We randomly divide our overall sample into k subsets or folds.
- We train our algorithm on k - 1 folds, holding just one group out for model assessment.
- We repeat this process k times—every fold is held out once and used to fit the model k - 1 times.
- We then pool or average the evaluation metrics (e.g., predictive accuracy) for all the held-out runs.
Stratified k-fold cross-validation ensures that the distribution of class labels (or for numeric targets, the mean) is relatively constant across folds.
Hyperparameter Optimization
In SML settings, we automatically learn the parameters (e.g., coefficients) of our algorithms during estimation.
Hyperparameters, on the other hand, are chosen by the analyst, guide the entire learning or estimation process, and can powerfully shape our algorithm’s predictive performance.
- Examples of hyperparameters include the k in nearest neighbours algorithms, the \alpha penalty term in ridge regressions, or the number of hidden layers in a neural network.
How can analysts settle on the right hyperparameter value(s) for their algorithm?
- Test different values via trial and error.
- Use automated procedures like
GridSearchCVfromscikit-learn.
k-Nearest Neighbours
Brief Overview of KNN
k-nearest neighbours (KNNs) are simple, non-parametric algorithms that predict values of Y based on the distance between rows (or observations’ inputs).
The estimation of KNNs proceeds as follows:
- The analyst defines the distance metric (e.g., Euclidean, Manhattan) they will use to determine how similar any two observations are based on their vector of features.
- The analyst defines a value for the hyperparameter k—that is, the number of nearest neighbours to find in the training subsample.
- When fed a new data point, KNNs find the k nearest neighbours in the training data, and:
- Assign the new observation to the class that most of its k nearest neighbours belong to (for classification problems.)
- Generate a prediction of Y by taking the average of the target variable for the new observation’s k nearest neighbours (for regression problems.)
- Assign the new observation to the class that most of its k nearest neighbours belong to (for classification problems.)
KNN in Python
Initial Steps
# Importing pandas to wrangle the data, modules from scikit-learn to fit KNN classifier,
# and numpy to iterate over potential values of k (for a grid search):
import pandas as pd
from sklearn.model_selection import train_test_split, cross_val_score, StratifiedKFold, GridSearchCV
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KNeighborsClassifier
import numpy as np
# Load input data frame:
gapminder = pd.read_excel("https://github.com/sakeefkarim/intro.python.24/raw/main/data/gapminder.xlsx")
# Homing-in on observations in the latest year (2007)
gapminder = gapminder.query("year == 2007").reset_index(drop=True).drop(columns='year')
# Generating dummy indicator indexing whether a country is in Asia:
gapminder['asia'] = pd.get_dummies(gapminder['continent'])['Asia']
# Removing target variable and categorical indicators from feature vector:
X = gapminder.drop(columns = ['asia', 'continent', 'country'])
# Isolating target variable:
y = gapminder['asia']Train-Test Split, Cross-Validation
# Perform train-test split:
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify = y, test_size = 0.15, random_state = 905)
# Standardizing feature data
sc = StandardScaler()
# Applied to training data
X_train = sc.fit_transform(X_train)
# Applied to test data
X_test = sc.transform(X_test)
# Initializing KNN classifier with k = 5, fitting model:
knn = KNeighborsClassifier(n_neighbors = 5).fit(X_train, y_train)
# Stratified k-fold cross-validation:
skfold = StratifiedKFold(n_splits = 5, shuffle = True, random_state = 905)
# Cross-validation score:
knn_5_cv = cross_val_score(knn, X_train, y_train, cv = skfold).mean()
# Measure of predictive performance:
knn_5_test = knn.score(X_test, y_test)
print(f"Cross-Validation Score: {knn_5_cv:.3f},\nTest Set Score: {knn_5_test:.3f}")Cross-Validation Score: 0.792,
Test Set Score: 0.818Hyperparameter Optimization
# Creating a grid of potential hyperparameter values (odd numbers from 1 to 13):
k_grid = {'n_neighbors': np.arange(start = 1, stop = 14, step = 2) }
# Setting up a grid search to home-in on best value of k:
grid = GridSearchCV(KNeighborsClassifier(), param_grid = k_grid, cv = skfold)
grid_fit = grid.fit(X_train, y_train)
print(f"Best Mean Cross-Validation Score: {grid.best_score_:.3f},\nBest Parameters (Value of k): {grid.best_params_},\nTest Set Score: {grid.score(X_test, y_test):.3f}")Best Mean Cross-Validation Score: 0.817,
Best Parameters (Value of k): {'n_neighbors': 3},
Test Set Score: 0.773Note: Keep clicking or the space bar on your to advance through this slide.
Memo II
Five Survey Items
Please use the rest of today’s session to
work on your second response memo!
Enjoy the Weekend
References
Note: Scroll to access the entire bibliography